Tempus xE - Whole Exome Next-Generation Sequencing Assay
Tempus xE (version 2) is a whole exome next-generation sequencing assay that analyzes the entire coding region (exome) of the patient’s genome, combined with whole transcriptome RNA sequencing. Clinical sequencing is performed to 500x depth of coverage for tumor specimens and 150x for normal specimens for the clinically enhanced regions (648 genes). Non-enhanced regions are performed at 250x depth of coverage for tumor specimens and 150x for normal specimens.
It encompasses 19,433 genes covering a ~34 Mb target region of the human genome and is optimized for formalin fixed paraffin embedded (FFPE) tissue samples. The FFPE tumor tissue is matched to a normal blood or saliva sample to ensure fidelity of somatic variant calling. The xE assay identifies actionable oncogenic variants. From DNA sequencing, somatic and incidentally detected germline single nucleotide variants (SNVs), insertions and deletions (indels), and copy number gains (CNGs). From RNA-seq, gene fusions (translocations) are detected in an unbiased and comprehensive manner. In addition, tumor mutational burden (TMB) is reported. Microsatellite Instability (MSI) status is available as a research use only metric upon request. Whole transcriptome RNA-seq is 50 million reads.
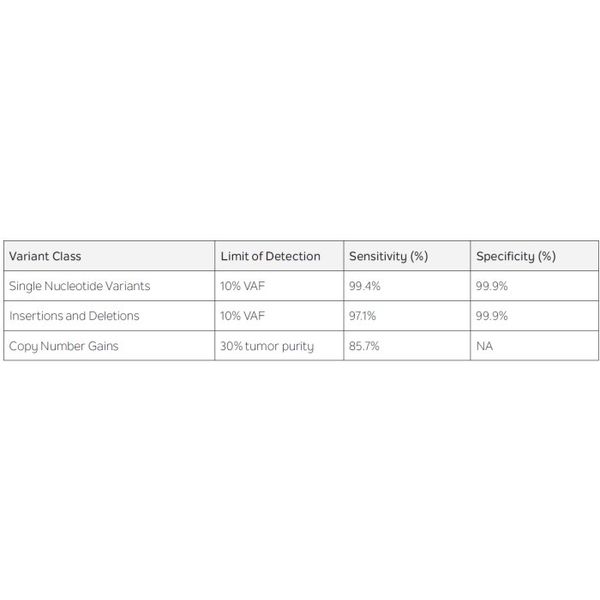
CAP/CLIA validation of the Tempus xE (version 2) assay focused on actionable oncologic variants. The assay requires specimens with tumor content of at least 40% post macrodissection. Performance specifications are listed in Table 1 below as compared to internally developed orthogonal tests. These results establish high sensitivity and specificity for the Tempus xE (version 2) assay.