

- Home
- Companies
- SOPHiA Genetics
- Software
- MSK-ACCESS Powered with SOPHiA DDM - ...
MSK-ACCESS Powered with SOPHiA DDM - In-House Liquid Biopsy
Rise above the noise to see what truly matters. Elevate your precision oncology program. Push the boundaries of your in-house liquid biopsy capabilities with MSK-ACCESS® powered with SOPHiA DDM™, a decentralized version of the rigorously validated cell-free DNA (cfDNA) assay developed and used in clinical routine by Memorial Sloan Kettering Cancer Center (MSK)1,2. Discover an innovative solution that combines MSK’s expertise in cancer genomics with the robust analytics of the SOPHiA DDM™ Platform for paradigm-shifting liquid biopsy insights.
Clinical knowledge-driven design
Focus on what matters with a comprehensive 146 gene panel curated by MSK experts.
End-to-end application
Save time with a streamlined workflow, taking you from cfDNA sample to report in ∼5 days.
Matched tumor-normal approach
Reduce noise by filtering clonal hematopoiesis of indeterminate potential (CHIP) and germline variants.
Robust analytical performance
Trust your results with demonstrated 99.4% positive percent agreement (≥0.5% VAF) to MSK-ACCESS® used at MSK3.
Tumor-informed variant calling
Maximize insights with informed variant calling across sequential tests, facilitating longitudinal monitoring.
Integrated analysis and reporting
Increase efficiency with the robust analytics and user-friendly interpretation and reporting features of SOPHiA DDM™.
Equip your laboratory with a true sample-to-report workflow that combines in-house hybrid capture and cloud-based analytics, allowing you to to retain ownership of your samples and data.
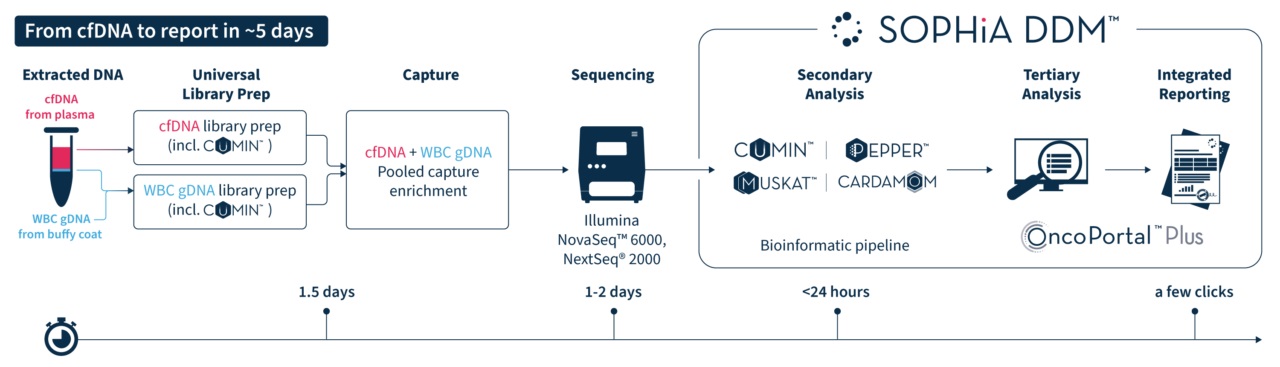
- 146 gene panel focused on actionability, curated by MSK experts and selected from MSK’s cancer genomic profiling assay, MSK-IMPACT®
- Hybridization-based capture with deep sequencing (~20,000x)
- Tailored probes for high on-target rate and coverage uniformity
- Minimal input amount of only 20 ng cfDNA
- Ready-to-sequence libraries in just 1.5 days
- Optimized multiplexing of paired tumor-normal samples for a cost-effective process
- Compatible with NovaSeq™ 6000 and NextSeq® 2000 sequencers
- Algorithm-powered detection of SNVs, Indels, CNV, and fusions
- Proprietary unique molecular identifier (UMI) technology, CUMIN™, for sensitive variant detection down to 0.5% VAF
- Tumor-informed variant calling to facilitate follow-ups and decision-making
- Tertiary analysis based on the latest scientific evidence on relevant variants with OncoPortal™ Plus
- Access to MSK’s Precision Oncology Knowledge Base, OncoKB™, via link-out at gene level for enhanced interpretation support
- Knowledge-sharing within SOPHiA GENETICS Community, one of the largest networks of connected healthcare institutions
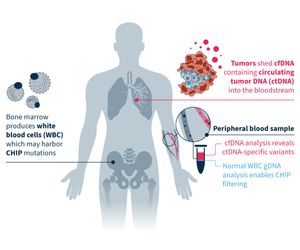
Clonal hematopoiesis of indeterminate potential (CHIP) is a common cause of biological false positives in cfDNA analysis4. MSK-ACCESS® powered with SOPHiA DDM™ leverages a matched tumor-normal approach that enables CHIP filtering and eliminates the risk of:
- Biological false positives at low allele frequencies (e.g. in BRCA1).
- Removing true positives, if using only database-driven approaches to annotate CHIP2.
By sequencing tumor-derived cfDNA and normal white blood cell DNA in parallel, you can effectively reduce noise in your data analysis and focus on tumor-specific somatic variants.
- Content: 146 genes
- Diseases Covered: Multi-cancer (any solid tumor)
- Detected Variants
- SNVs and Indels in 146 genes
- CNVs in 71 genes
- Fusions (DNA-based via intronic targets) in 10 genes
- TERT promoter
- Sample Type: Paired samples of cell-free DNA extracted from peripheral blood and white blood cell gDNA
- Starting Material: 20 ng cell-free DNA (minimum); 50 ng white blood cell (WBC) gDNA (minimum)
- Recommended Read Length: 150 bp, paired end
- Coverage Depth: Approx. 20,000x
- Multiplexing (samples per run)
- Illumina NovaSeq™ 6000:
- SP flow cell: 8 (tumor + normal) pairs
- S1 flow cell: 16 (tumor + normal) pairs
- S2 flow cell: 40 (tumor + normal) pairs
- S4 flow cell: 96 (tumor + normal) pairs
- Illumina NextSeq® 2000 (P3 flow cell):
- 8 (tumor + normal) pairs, with 33% sequencing space left
- Library Preparation Time: 1.5 days
