VeraChem, LLC
- Home
- Companies
- VeraChem, LLC
- Software
- VeraChem - Version VMap 0.9 - Small ...
VeraChem - Model VMap 0.9 - Small Molecule Tools
FromVeraChem, LLC
Superimpose related ligand series on reference structure Reference can be co-xtal PDB, sdf, mol, crd Handles arbitrary atom orderings. Provides excellent VM2/docking starting structures. VMap is an application to superimpose related ligands (e.g. common scaffold atoms; different R-groups) on a reference molecule. The reference can be, for example, a co-crystalized ligand taken from a protein PDB file, in which case, VMap provides excellent starting structures for VM2 or docking calculations.
- Superimpose congeneric ligand series on a reference structure
Identifies the maximum common substructure (MCS) shared by the reference molecule and each of a series of ligands. This information is then used to superimpose each ligand in the series on the reference molecule, maximizing overlap by dihedral rotations. - Multiple formats supported
Reference format can be PDB (e.g. co-xtal ligand), SDF, MOL, or CRD. Ligand series supplied as SDF. - Handles arbitrary atom numbering/ordering
Equivalent atoms mapped automatically – no atom reordering required. - 1-Click interface with VDisplay molecular visualization of results
Send superimposed structures to Verachem’s VDisplay. Output of atom mappings of the maximum common substructures in a CSV file.
VMap seamlessly interfaces with VDisplay to visualize the superimposed molecular structures produced.
Vmap version 0.9 for Microsoft Windows®.
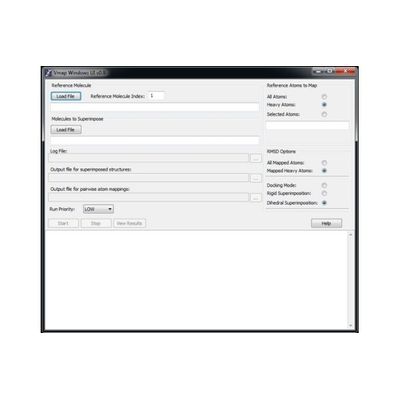
The VMap graphical user interface provides detailed user control, including:
- can select molecule is reference from a multi-molecule file
- if all-atom reference choose wether to include hydrogen atoms in mapping all atom or just heavy-atoms
- choose whether RMSD calculation includes hydrogen atoms or just heavy atoms
- choose from rigid superposition or overlap maximization via dihedral rotations