- Home
- Companies
- Claret Bioscience
- Software
- Claret - Srsly UMI Post Processing ...
Claret - Srsly UMI Post Processing Software
Though there are many ways to handle UMI sequences with reads, we show only one example on this page. The SRSLY UMI software tool will demultiplex samples with bcl2fastq and place UMI sequences in the FASTQ headerline for each fragment. After mapping FASTQ into BAM format, the UMI sequence can then be moved to an auxiliary read tag (such as RX: or BC:) for further analysis steps. We provide SRSLYumi, a simple Python package for running BCL2FASTQ, as well as describing the steps for a manual configuration.
1. Install SRSLYumi with:
pip install srslyumi
The package is compatible with Python 2.7 and 3, and can be installed in a virtual environment if necessary. You should also have bcl2fastq installed on your system.
Alternatively, manually download the SRSLYumi code from GitHub
2. After sequencing, identify the location of the Illumina run directory that contains both a RunInfo.xml, SampleSheet.csv, and the subdirectory /Data/Intensities/BaseCalls. This is the “run directory” and the first parameter to the script. If these are not located in the same directory, their locations can be specified separately.
3. Run:
srslyumi [rundirectory] [outputdirectory]
FASTQ will be produced in [outputdirectory].
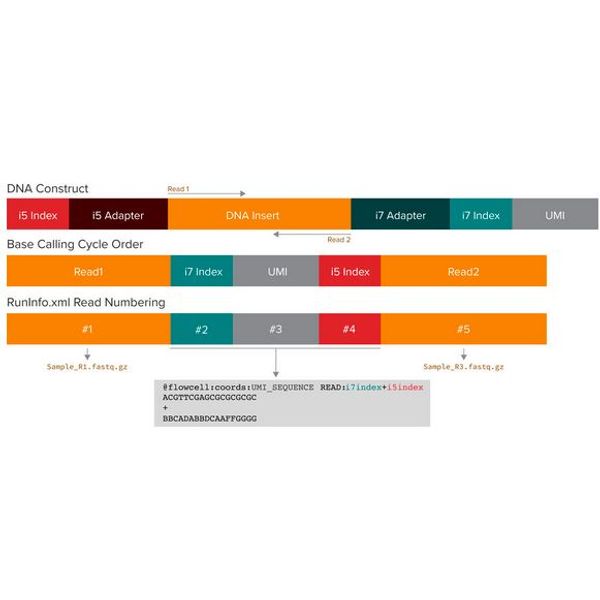
UMI-aware demultiplexing (Manual)Manual demultiplexing requires additional configuration of the BCL2FASTQ run. The read cycles for the UMI occur in between the read cycles for the two indices. BCL2FASTQ can automatically take the first base pairs of read 2 and place them in the FASTQ header as UMIs. So instead of the usual 4 bcl2fastq reads (read1, index1, index2, read2), we set up 5 reads:
-
Read 1 (the normal R1 FASTQ file)
-
Index 1 (the normal index read)
-
Read 2 (actually the UMI, will be placed into the FASTQ header)
-
Index 2 (the normal index read)
-
Read 3 (actually the normal R2 FASTQ file)
1. Thesection of RunInfo.xml must be edited to something like the following, where NumCycles depends on the read length in the run (e.g. your 151 may be 101):
2. The SampleSheet.csv [Reads] section should specify the non-indexed reads as in the RunInfo.xml, for example:
[Reads],,,,,,,,,, 151,,,,,,,,,, 9,,,,,,,,,, 151,,,,,,,,,, ,,,,,,,,,,
3. The SampleSheet.csv must have TrimUMI, Read2UMIStartFromCycle, and Read2UMILength in the [Setting] section:
[Settings],,,,,,,,, ReverseComplement,0,,,,,,,, TrimUMI,1,,,,,,,, Read2UMIStartFromCycle,1,,,,,,,, Read2UMILength,9,,,,,,,,
In SampleSheet.csv, the Index and Index2 sequences must be 8 bp long.
For information about sequencer specific set-up for UMI-aware run contact Illumina® technical support.